Flourescence intensities in plant roots¶
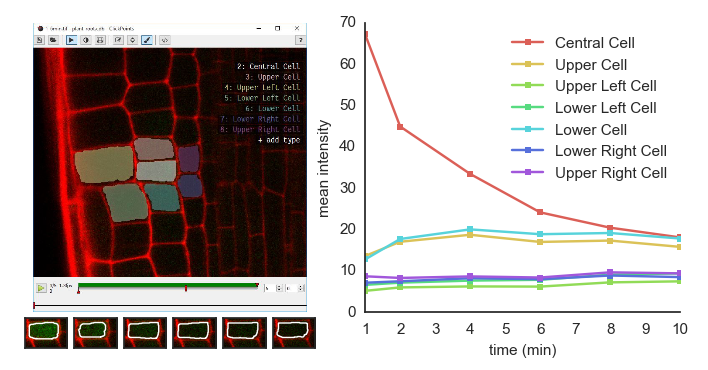
Left: image of a plant root in ClickPoints. Right: fluorescence intensities of the cells over time.
In the example, we show how the mask panting feature of ClickPoints can be used to evaluate fluorescence intensities in microscope recordings.
Images of an Arabidopsis thaliana root tip, obtained using a two-photon confocal microscope [1], recorded at 1 min time intervals are used. The plant roots expressed a photoactivatable green fluorescent protein, which after activation with a UV pulse diffuses from the activated cells to the neighbouring cells.
For each time step a mask is painted to cover each cell in each time step.
The fluorescence intensities be evaluated using a small script:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 | from __future__ import division, print_function
import re
import numpy as np
from matplotlib import pyplot as plt
# connect to ClickPoints database
# database filename is supplied as command line argument when started from ClickPoints
import clickpoints
start_frame, database, port = clickpoints.GetCommandLineArgs()
db = clickpoints.DataFile(database)
com = clickpoints.Commands(port, catch_terminate_signal=True)
# get images and mask_types
images = db.getImages()
mask_types = db.getMaskTypes()
# regular expression to get time from filename
regex = re.compile(r".*(?P<experiment>\d*)-(?P<time>\d*)min")
# initialize arrays for times and intensities
times = []
intensities = []
# iterate over all images
for image in images:
print("Image", image.filename)
# get time from filename
time = float(regex.match(image.filename).groupdict()["time"])
times.append(time)
# get mask and green channel of image
mask = image.mask.data
green_channel = image.data[:, :, 1]
# sum the pixel intensities for every channel
intensities.append([np.mean(green_channel[mask == mask_type.index]) for mask_type in mask_types])
# convert lists to numpy arrays
intensities = np.array(intensities).T
times = np.array(times)
# iterate over cells
for mask_type, cell_int in zip(mask_types, intensities):
plt.plot(times, cell_int, "-s", label=mask_type.name)
# add legend and labels
plt.legend()
plt.xlabel("time (min)")
plt.ylabel("mean intensity")
# display the plot
plt.show()
|
References
| [1] | Nadja Gerlitz. Dronpa. 2016. |